Integrating Machine Learning with Total Network Controllability Analysis to Identify Therapeutic Targets for Cancer Treatment
Other Articles
By analysing huge amounts of biological data, the use of machine learning accelerates the identification of critical control hubs that are sensitive to changes in the network structure of the total controllability network, thereby having potential as diagnostic biomarkers and therapeutic targets for disease and cancer treatment.
Study conducted by Prof. Weixiong ZHANG and his research team
Mutations in genes are the primary cause of cancer. Cancer research has mainly focused on identifying cancer-driver genes (CDGs) that may trigger tumorigenesis or promote aberrant cell growth. Modern large-scale sequencing of human cancers aims to comprehensively discover mutated genes that confer a selective advantage to cancer cells1. However, there is a lack of a widely accepted gold standard for CDGs, as cancer is highly heterogeneous, and different cancers are driven by distinct sets of genetic mutations.
In a study published in iScience, a research team led by Weixiong ZHANG, Associate Director of PolyU Academy for Interdisciplinary Research (PAIR) and Chair Professor of Bioinformatics and Integrative Genomics in the Department of Health Technology and Informatics at the Hong Kong Polytechnic University, Hong Kong Global STEM Scholar, took a different approach, in which they identify genes that maintain cancerous cell states, which they termed “cancer-keeper genes (CKGs)”2. Unlike driver genes, whose mutations directly contribute to cancer initiation and progression, keeper genes are essential for maintaining cellular homeostasis and survival. Interventions targeting CKGs may terminate or prevent aberrant cell differentiation and proliferation, making them ideal biomarkers for diagnosis and therapeutic targets.
With the aid of machine learning in developing a gene regulatory network (GRN), the research team extended the theory of total network controllability and developed an efficient algorithm to identify CKGs. The concept is grounded in control theory and is particularly relevant in systems represented by graphs, where nodes represent entities and edges represent interactions. A network is considered totally controllable if it is possible to manipulate the states of all nodes using a finite set of control inputs applied to specific nodes. It has been used in electrical engineering to characterise power grids and transportation networks. In the context of biological systems, this analysis helps identify key components, or “control hubs”, which are crucial to influencing the behaviour of the entire network, making them ideal candidates for therapeutic interventions (Figure 1a). The research team constructed a GRN on protein interaction data and signalling pathway information describing regulatory relationships among genes (Figure 2). The network consists of cancer-related genes (as seed nodes) and edges capturing their interactions to transverse the ten important signalling pathways selected from five well-curated, disease- and cancer-related pathway databases.
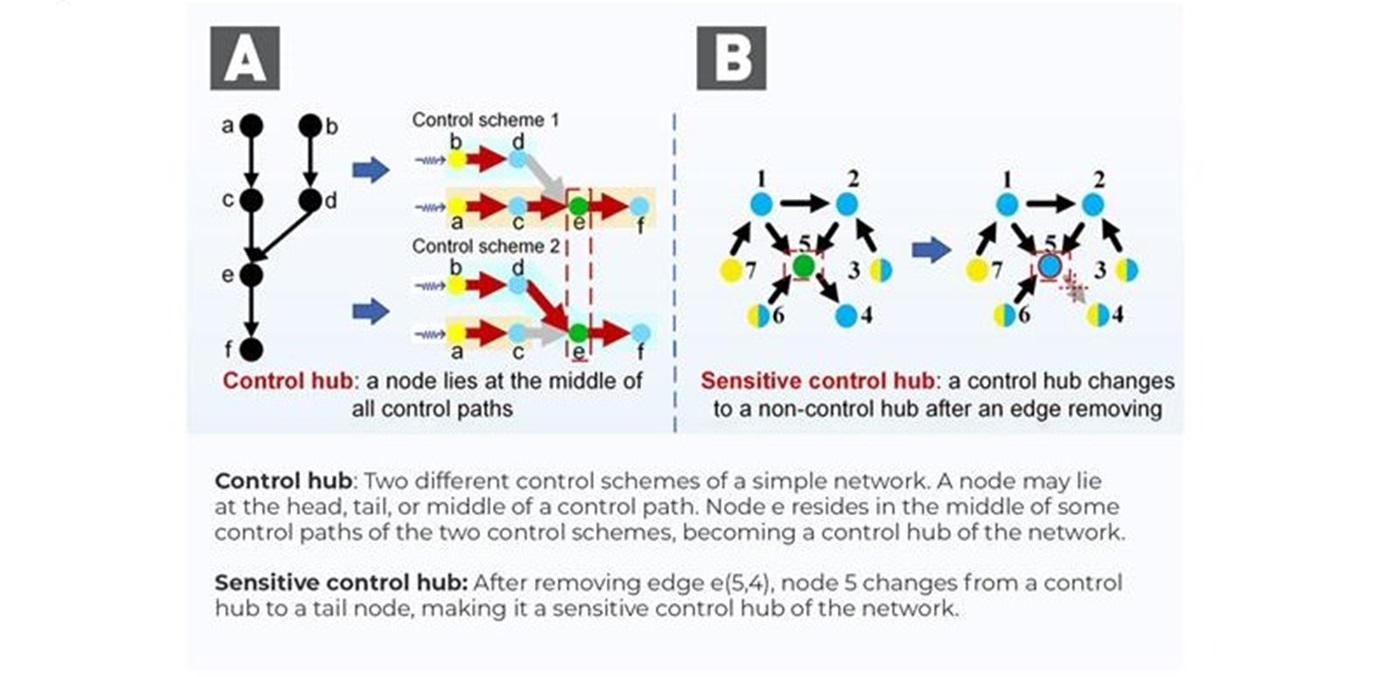
Figure 1. An illustration of control hubs and sensitive control hubs

Figure 2. Identification of CKGs and sCKGs in Cancer Gene Regulatory Network (GRN)
In the study, the research team considered control hubs candidates for abnormal cellular CKG, noting that some control hubs could be more sensitive and vulnerable to external perturbations than others. They focused on those control hubs that could be turned into non-control hubs when a single edge is removed from the network as a form of perturbation (Figure 1b). Such sensitive CKGs (sCKGs) are considered better therapeutic targets.
Machine learning techniques are applied to explore vast amounts of genetic data to construct biological networks and identify patterns and relationships in the networks that may not be immediately obvious. A novel polynomial-time algorithm was developed to identify all control hubs without the need to compute all control schemes of a network3. The algorithm first identifies the head and tail nodes of the control paths of all control schemes and subsequently identifies the control hubs. This analysis helps identify the nodes in a network that are crucial for controlling the system’s behaviour, making them suitable candidates for therapeutic targets.
The research team applied the CKG approach and constructed a GRN for bladder cancer (BLCA), which consists of 7,030 nodes (genes) and 103,360 directed edges. By a machine learning approach, 660 nodes were identified as control hubs (CKGs), of which only 173 nodes were classified as sCKGs. When mapping with a network that illustrates the interactions between proteins within human cells, 35 sCKGs were considered potential therapeutic targets (Figure 2). Remarkably, all genes involved in the cell-cycle and p53 pathways in BLCA were identified as CKGs. Experiments on cell lines and a mouse model confirmed that six sensitive CKGs effectively suppressed cancer cell growth (Figure 3).
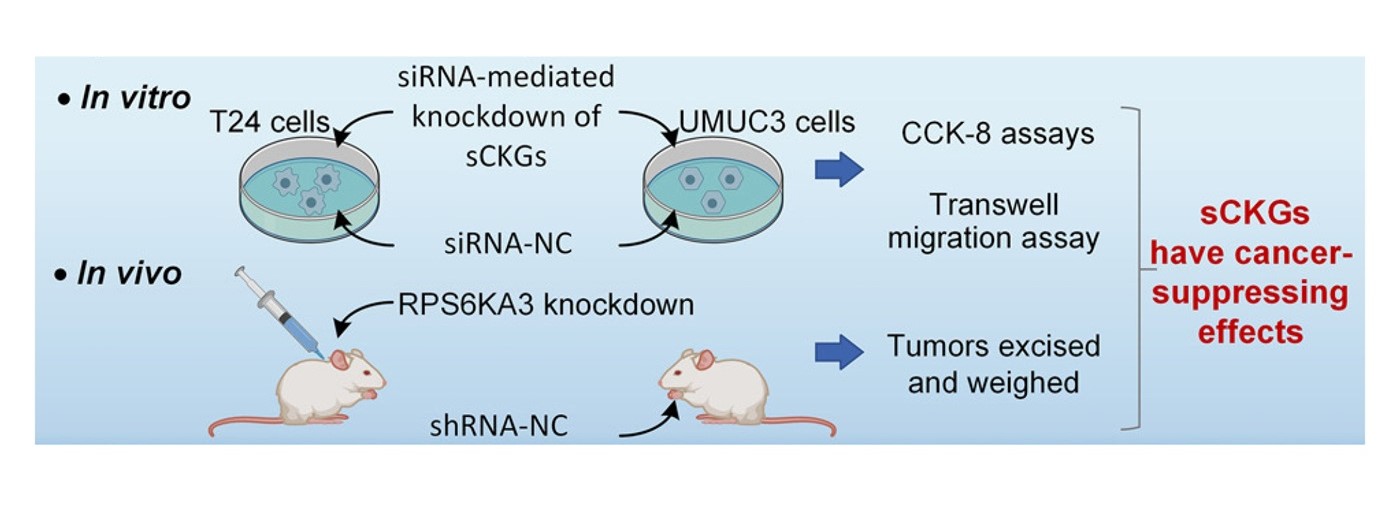
Figure 3. Experimental validation results in cell lines and a mouse cancer model
The regulatory network constructed in the study is a pan-cancer gene regulatory network suitable for applying network controllability. In addition to using seed genes specific to one type of cancer, the network could be modified to target another by removing incompatible genes and interactions detected under different conditions. The method using total network controllability analysis could also be extended to identify the control hubs of other diseases, for example, the SARS-CoV-2 infectious disease4.
The research was partly supported by the National Natural Science Foundation of China (grant numbers 62176129 and 81672523), the Scientific Research Foundation of Liaoning Province (LJKZ1134), the Hong Kong Global STEM Professorship Scheme, the Hong Kong Jockey Club Charities Trust, and a grant from the Hong Kong Health and Medical Fund (grant number 10211696).
The data for establishing gene regulatory networks in this study have been deposited at Github: https://github.com/network-control-lab/control-hubs. Additionally, in vivo experimental data from the mouse model, as reported in the study, are available upon request from the lead contact. All original code for finding control hubs and cancer-keeper genes are freely available at GitHub: https://github.com/network-control-lab/control-hubs.
| References |
|---|
1. Tokheim, C.J., Papadopoulos, N., Kinzler, K.W., Vogelstein, B., Karchin, R. Evaluating the evaluation of cancer driver genes. Proc Natl Acad Sci U S A. 2016 Dec 13;113(50):14330-14335. doi: 10.1073/pnas.1616440113.
2. Zhang, X., Pan, C., Wei, X., Yu, M., Liu, S., An, J., Yang, J., Wei, B., Hao, W., Yao, Y., Zhu, Y., Zhang, W. Cancer-keeper genes as therapeutic targets. iScience. 2023 Jul 11;26(8):107296. doi: 10.1016/j. isci.2023.107296.
3. Zhang, X., Pan, C., and Zhang, W. Control Hubs of Complex Networks and a Polynomial-Time Identification Algorithm. arXiv:2206.01188 (2022).
4. Wei, X., Pan, C., Zhang, X., … and Zhang, W. Total network controllability analysis discovers explainable drugs for Covid-19 treatment. Biol Direct 18, 55 (2023). https://doi.org/10.1186/s13062-023-00410-9
 | Prof. Weixiong ZHANG of PolyU Academy for Interdisciplinary Research (PAIR) and Integrative Genomics in the Department of Health Technology and Informatics, Hong Kong Global STEM Scholar |